Kedro for ML industrialization
In the following article, i will show how to install and use Kedro, an open source Python library that will simplify and clarify the way to define data pipelines for your projects.
Then we will create a project and a custom kedro dataset
Install kedro
First create a new conda virtual environment : kedro is compatible with python 3.6, 3.7 and 3.8 versions. Here i choosed 3.7.
conda create -n kedro_project python=3.7
Then activate it
conda activate kedro_project
Install kedro from conda-forge repository:
conda install -c conda-forge kedro
Install also kedro-viz plugin, very usefull to display your data pipelines:
Install kedro-viz plugin
pip install kedro-viz
Verify the installation:
kedro info
_ _
| | _____ __| |_ __ ___
| |/ / _ \/ _` | '__/ _ \
| < __/ (_| | | | (_) |
|_|\_\___|\__,_|_| \___/
v0.16.2
kedro allows teams to create analytics
projects. It is developed as part of
the Kedro initiative at QuantumBlack.
Installed plugins:
kedro_viz: 3.4.0 (hooks:global,line_magic)
Create your Data Science project directory
We are going to create our datascience project directory using Kedro. It will initialize all the needed directories using a cookiecutter template.
First go to the directory where you’re used to create your projects repos ( mine is /home/tdenimal/Dev )
cd /home/tdenimal/Dev
Then create project and choose the project name, leave the other choices at their default value. I choosed pneumothorax, to create the project directory for the following work.
kedro new
Project Name:
=============
Please enter a human readable name for your new project.
Spaces and punctuation are allowed.
[New Kedro Project]: pneumothorax
Repository Name:
================
Please enter a directory name for your new project repository.
Alphanumeric characters, hyphens and underscores are allowed.
Lowercase is recommended.
[pneumothorax]:
Python Package Name:
====================
Please enter a valid Python package name for your project package.
Alphanumeric characters and underscores are allowed.
Lowercase is recommended. Package name must start with a letter or underscore.
[pneumothorax]:
Generate Example Pipeline:
==========================
Do you want to generate an example pipeline in your project?
Good for first-time users. (default=N)
[y/N]:
Change directory to the project generated in /home/tdenimal/Dev/pneumothorax
A best-practice setup includes initialising git and creating a virtual environment before running `kedro install` to install project-specific dependencies. Refer to the Kedro documentation: https://kedro.readthedocs.io/
GIT init
As a best practice, you should add your newly created project directory to git right after creation:
cd pneumothorax
git init
git add --all
git commit
If you are a github user, you can create a repository with the same name on github and then push your local git.
First, you will need to add an ssh public key to your github profile.
You can generate a ssh key pair using
ssh-keygen
Then retrieve you public key from default location:
cat ~/.ssh/id_rsa.pub
Copy/paste the content of the .pub file to your github profile
Using this method, you will no longer need to submit your password each time you push your code to github.
Then push your code to github:
Just replace username with your github username.
git remote add origin git@github.com:username/${PWD##*/}
git push -u origin master
Defining Data pipeline
Install Dataset Dependencies
Our pipeline is using DICOMDataSet, to be able to read DICOM files and extract ImageDataSet and CSVDataSet from it. DICOMDataSet is a custom Kedro DataSet, you can see how it was created in the following article.
Kedro libraries dependencies are modularized, you can choose to install dependencies for a specic dataset.
For example, to install dependencies to be able to use the CSVDataSet:
pip install "kedro[pandas.CSVDataSet]"
Same thing for ImageDataSet which is using pillow library:
pip install "kedro[pillow.ImageDataSet]"
Kedro Data Catalog
Before being able to used Dataset in Kedro you have to define them in the Kedro Data Catalog which regroups all datasets in our project.
You can configure it using conf/base/catalog.yml file.
By default, Kedro Data Catalog follows a Data Engineering Convention to classify datasets and managing data. From raw (our initials datasets) to model input (ready to be used in our models).
It s a good practice to manage your datasets and group them logically.
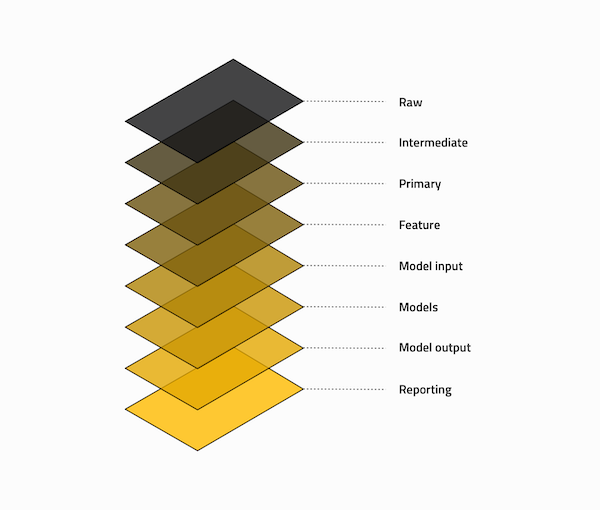
You are free to follow this scheme or define yours. I decided to follow the default one as it suits my needs well.
Here’s how Kedro team defines each layer :
| Folder in data | Description |
|---|---|
| Raw | Initial start of the pipeline, containing the sourced data model(s) that should never be changed, it forms your single source of truth to work from. These data models are typically un-typed in most cases e.g. csv, but this will vary from case to case. |
| Intermediate | Optional data model(s), which are introduced to type your raw data model(s), e.g.
converting string based values into their current typed representation. |
| Primary | Domain specific data model(s) containing cleansed, transformed and wrangled data from either
raw or intermediate, which forms your layer that you input into your feature
engineering. |
| Feature | Analytics specific data model(s) containing a set of features defined against the primary
data, which are grouped by feature area of analysis and stored against a common dimension. |
| Model input | Analytics specific data model(s) containing all feature data against a common dimension
and in the case of live projects against an analytics run date to ensure that you track the
historical changes of the features over time. |
| Models | Stored, serialised pre-trained machine learning models. |
| Model output | Analytics specific data model(s) containing the results generated by the model based on the
model input data. |
| Reporting | Reporting data model(s) that are used to combine a set of primary, feature,
model input and model output data used to drive the dashboard and the views
constructed. It encapsulates and removes the need to define any blending or joining of data,
improve performance and replacement of presentation layer without having to redefine the data
models. |
Data engineering pipeline
Step 1 - From .dcm files (raw) to .csv/.png files (intermediate/primary)
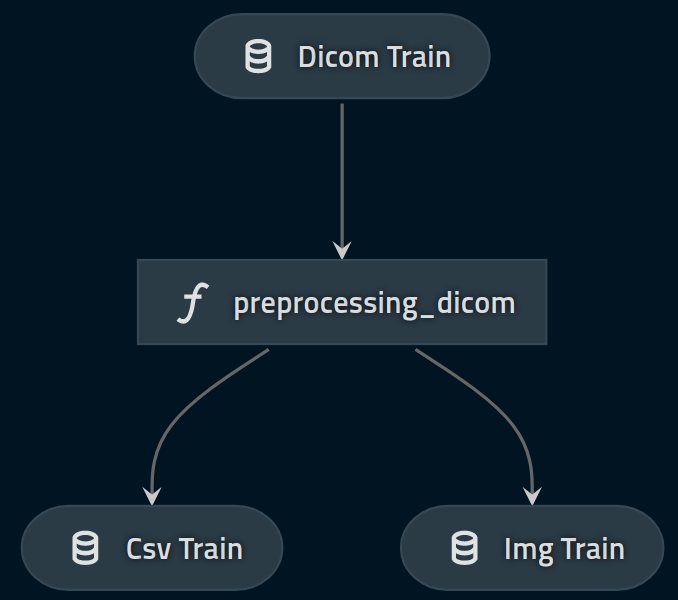
We are going to define the first part of our data engineering pipeline, working with dcm files 4, our raw dataset, to extract csv and png files.
Csv file is just an extract and concatenation of all metadata contained in dcm files. We will store it as intermediate dataset, as it is just a typing of our raw data.
The png files are extracted from dcm files but also resized for practical issues. We will store it as model input as it can be used directly without further modifications in our model.
Defining raw dataset
In the conf/base/catalog.yml file, we will parameter our raw dataset dicom_train, the train directory containing dcm files.
dicom_train:
type: PartitionedDataSet
dataset: pneumothorax.io.datasets.dicom_dataset.DICOMDataSet
path: data/01_raw/dicom-images-train
filename_suffix: ".dcm"
layer: raw
dicom_train is typed as PartitionedDataSet as we are going to read the raw directory as a whole.
Defining target datasets
In the conf/base/catalog.yml file, we will parameter our target datasets :
- csv_train : a csv file containing concatenated dcm metadata
- img_train : a directory to store .png files, extracted also from dcm files.
img_train:
type: PartitionedDataSet
dataset: pillow.ImageDataSet
path: data/02_intermediate/img_train
filename_suffix: ".png"
layer: intermediate
csv_train:
type: pandas.CSVDataSet
filepath: data/02_intermediate/train.csv
layer: intermediate
Defining the node function
Next step we are going to define the node function that will make the link between our raw dataset and the target datasets.
-
dicom_train is of type DICOMDataSet
-
img_train should be of type pillow.Image
-
csv_train should be of type pandas.DataFrame
Add (if it does not exist) or edit the following file src/pneumothorax/pipelines/data_engineering/nodes.py file. This file will contain all the nodes functions for data_engineering pipeline.
Here’s the whole code of our node function, the function array_to_img is simply used to convert a numpy.ndarray to pillow.Image.
import pandas as pd
import numpy as np
from typing import List,Dict,Tuple
from PIL import Image
def array_to_img(array: np.ndarray,
size: Tuple[int, int] = (256,256)) -> Image:
"""Convert np.ndarray (pixel array) to PIL.Image and resizes eventually
Args:
array (np.ndarray): the input pixel array
size (Tuple[int, int], optional): (width,height) of returned PIL.Image. Defaults to (256,256).
Returns:
Image: PIL.Image as output
"""
return Image.fromarray(array).resize(size=(size[0], size[1]))
def preprocess_dicom(dicom: Dict) -> List:
"""Extract data from dicom.
Args:
dicom (Dict): dcm files ( PartitionedDataSet )
Returns:
List: [pandas.DataFrame (csv file), Dict of PIL.IMage ( PartitionedDataSet )
"""
#Init data with first dict item
imgs = {}
partition_id, partition_load_func = dicom.popitem()
partition_data = partition_load_func()
csv = partition_data[0]
imgs[partition_id] = array_to_img(partition_data[1])
#Loop on dict to extract img and csv data
for partition_id, partition_load_func in dicom.items():
partition_data = partition_load_func()
imgs[partition_id] = array_to_img(partition_data[1])
csv = pd.concat(
[csv, partition_data[0]], ignore_index=True, sort=True
)
return [csv,imgs]
In the fonction preprocess_dicom, the input dicom is of type Dict due to the way we load our dataset as a PartitionedDataSet, used to read a whole directory of file :
def preprocess_dicom(dicom: Dict) -> List:
The other specific part is when you retrieve an item from the Dict, in fact you retrieve a tuple containing the partition ID (in our case the filename) and the load() method bound to the underlying DICOMDataset object.
You can test this using the ipython shell embedded in Kedro :
kedro ipython
-------------------------------------------------------------------------------
Starting a Kedro session with the following variables in scope
startup_error, context
Use the line magic %reload_kedro to refresh them
or to see the error message if they are undefined
-------------------------------------------------------------------------------
ipython
Python 3.7.7 (default, May 7 2020, 21:25:33)
Type 'copyright', 'credits' or 'license' for more information
IPython 7.17.0 -- An enhanced Interactive Python. Type '?' for help.
2020-08-26 23:10:47,305 - root - INFO - ** Kedro project pneumothorax
2020-08-26 23:10:47,305 - root - INFO - Defined global variable `context` and `catalog`
2020-08-26 23:10:47,643 - root - INFO - Registered line magic `run_viz`
Then load dataset and let’s retrieve first item
In [1]: dicom_train = context.catalog.load("dicom_train")
2020-08-26 23:37:13,552 - kedro.io.data_catalog - INFO - Loading data from `dicom_train` (PartitionedDataSet)...
In [2]: type(dicom_train)
Out[2]: dict
In [3]: item = dicom_train.popitem()
In [4]: type(item)
Out[4]: tuple
In [5]: print(item[0])
1.2.276.0.7230010.3.1.4.8323329.7052.1517875202.524577
In [6]: print(item[1])
<bound method AbstractDataSet.load of <pneumothorax.io.datasets.dicom_dataset.DICOMDataSet object at 0x7f9471244dd0>>
In [7]: type(item[1]()[0])
Out[7]: pandas.core.frame.DataFrame
In [8]: type(item[1]()[1])
Out[8]: numpy.ndarray
In a nutshell we are going to loop on all dcm files contained in dicom_train, reading the output of the load() method of each DICOMDataset object.
DICOMDataset.load() output is a list containing the metadata info as a pandas DataFrame and a numpy.ndarray which is the pixel array of the xray image. All the metadata info will be concatenated in a single pandas DataFrame.
Defining the pipeline
Then we define our Kedro pipeline as a Kedro node that take dicom_train dataset as an input, calling preprocess_dicom method to output 2 news datasets, csv_train and img_train. You can observe that when a node function outputs more than 1 dataset, you define the outputs datasets in a List.
Add (if it does not exist) or edit the following file src/pneumothorax/pipelines/data_engineering/pipeline.py file.
from kedro.pipeline import node, Pipeline
from pneumothorax.pipelines.data_engineering.nodes import (
preprocess_dicom,
)
def create_pipeline(**kwargs):
return Pipeline(
[
node(
func=preprocess_dicom,
inputs="dicom_train",
outputs=["csv_train","img_train"],
name="preprocessing_dicom",
),
]
)
Add to master pipeline
Last step, add our Data engineering pipeline to the main pipeline.
Edit the src/pneumothorax/pipeline.py file.
"""Construction of the master pipeline.
"""
from typing import Dict
from kedro.pipeline import Pipeline
from pneumothorax.pipelines.data_engineering import pipeline as de
def create_pipelines(**kwargs) -> Dict[str, Pipeline]:
"""Create the project's pipeline.
Args:
kwargs: Ignore any additional arguments added in the future.
Returns:
A mapping from a pipeline name to a ``Pipeline`` object.
"""
de_pipeline = de.create_pipeline()
return {
"de": de_pipeline,
"__default__": de_pipeline,
}