Pneumothorax xray classifier - EDA - part 1 of 3
The goal of this project is to show a way how to train a binary classifier on Xray images, to predict if there is a pneumothorax or not. The dataset used here can be downloaded freely on Kaggle.
The project will be divided in 3 parts, in part 1 ( this one ), we will make an exploratory data analysis (EDA) to understand better the different datasets and what the target means ( Pneumothorax ). In part 2, we will define, train and test different binary classifier models. Last but not least, in part 3, we will implement Gradcam, which is a method to make computer vision algorithms results more interpretable.
This project code is available on Github and has been packaged using Kedro framework.
Target explanation - What is a Pneumothorax ?
Pneumothorax is an abnormal collection of air in the pleural space that causes an uncoupling of the lung from the chest wall. It can precipitate a life-threatening emergency due to lung collapse and respiratory or circulatory distress.
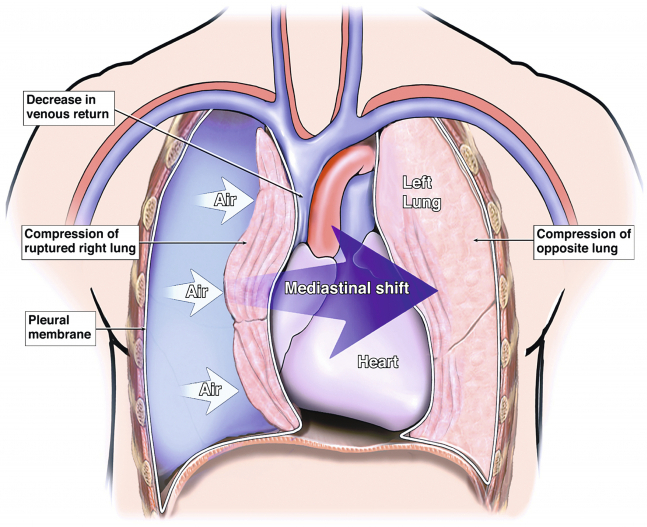
On some critical pneumothorax (tension pneumothorax), the region called mediastinum, containing heart,esophagus, the trachea especially, can be shifted to the other side, compressing the opposite lung. ( see the preceding infographic )
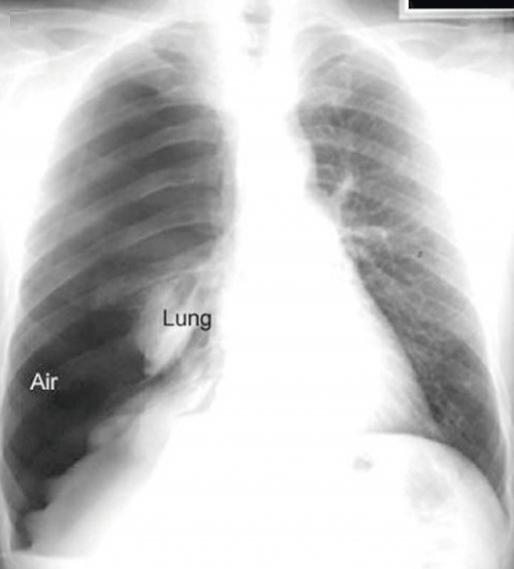
The preceding xray shows the same tension pneumothorax as before.
A pneumothorax can be caused by a blunt or penetrating chest injury, certain medical procedures, or damage from underlying lung disease.
Treatment for a pneumothorax usually involves inserting a needle or chest tube between the ribs to remove the excess air. However, a small pneumothorax may heal on its own.
For more information : Pneumothorax
Now that we know what we seek, it’s time to see what we have in our dataset.
DICOM Files
The dataset consists of DICOM (Digital imaging and communications in medicine) files, which is a standard file format for medical imaging information. See the definition in Wikipedia.
The pydicom python can be used to parse the DICOM files. There is also a csv file containing the pneumothorax zones by image, if present. The kaggle challenge was an image segmentation challenge but for the moment we will try to predict if an image contains or not a pneumothorax (binary classifier).
First, let’s see how much data we have in our dataset.
path_data_raw = "../data/raw"
path_data_cleaned = "../data/cleaned"
path_data_refined = "../data/refined"
metadata = pd.read_csv(path_data_raw+"/train-rle.csv")
metadata.info()
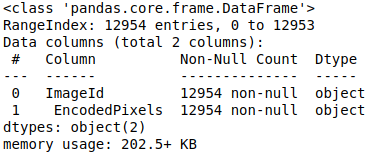
There is 12954 images in our dataset. Let’s see an extract of a csv file:

DICOM Files and metadata
Each dicom file contains a 1024*1024 jpg file.
The EncodedPixels contains a mask if there is 1 or several pneumothorax zones on xray. Otherwise it is set to -1 (No pneumothorax). There is much more metadata information in DICOM files. Let’s open one to see the content available.
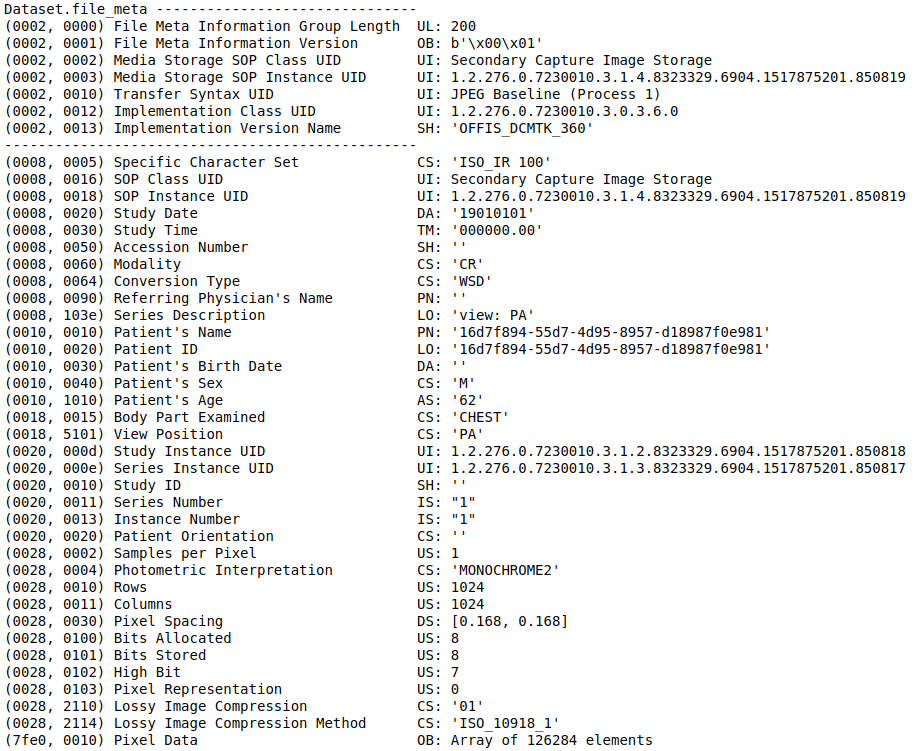
There is a lot of information in a DICOM file. We will select just a few to begin. An interesting one is the view position. AP stands for Anterior-Posterior, PA for Posterior-Anterior. It corresponds to the way the X-ray beam passing through the patient. See Wikipedia.
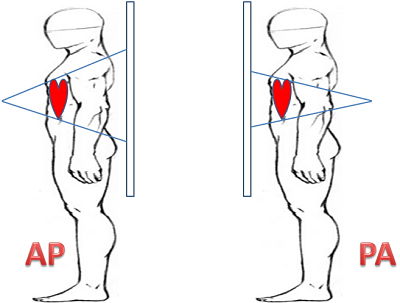
The xray result is quite different and it should be an interesting feature for our model.
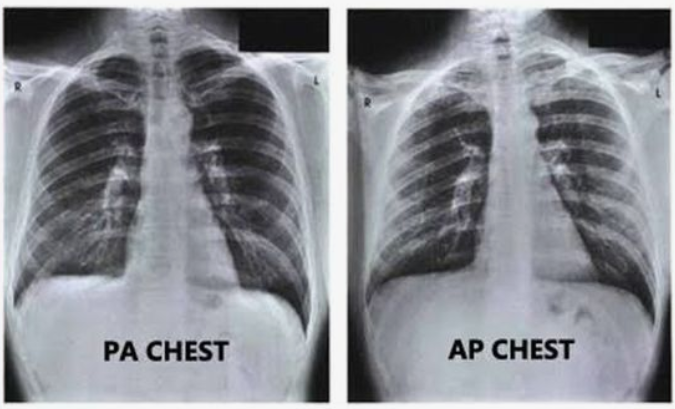
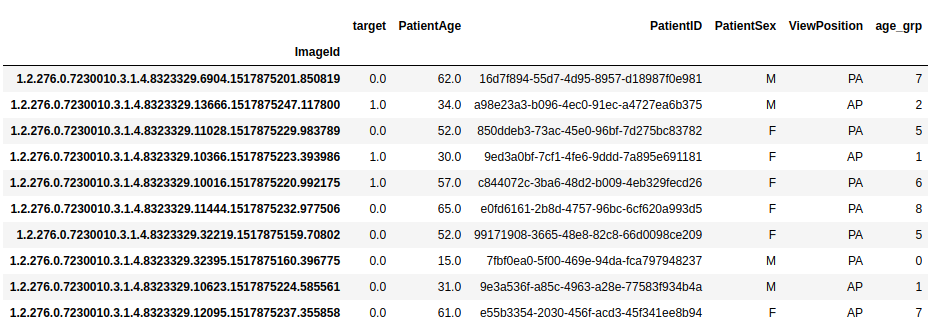
We will also select Patient Age, Patient Sex, which are obvious as features. Let’s add a function to read the metadata from our DICOM files.
def readMetaData(Dir, DF=pd.DataFrame()):
Files = glob(Dir+'*.dcm')
for file in Files:
ID = file.split('/')[-1][:-4]
DCM = pydicom.dcmread(file)
for atr in ['PatientAge', 'PatientID', 'PatientSex',"ViewPosition"]:
DF.loc[ID, atr] = getattr(DCM, atr)
return DF
trainDF = readMetaData(path_data_raw+"/dicom-images-train/", metadata)
trainDF.head(10)
Data cleaning, missing values, outliers
As we try to do only a binary classifier, just add a binary target column.
#Add binary target column
metadata["target"] = np.where(metadata[" EncodedPixels"] == "-1",0,1)
Now let’s check the class imbalance.
#Check Dataset imbalance
x=metadata["target"].value_counts().values
sns.barplot([0,1],x)
plt.title('Target variable count')
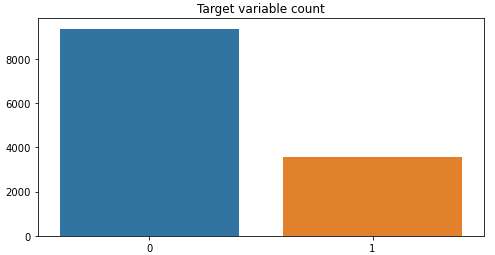
The dataset is imbalanced 1 pneumothorax xray for 2.62 normal xrays.
What next ?
In part 2, we will train classifier using different methods to obtain the desired accuracy.